By Giuliano Maciocci, Emmy Tsang, Nokome Bentley and Michael Aufreiter
In February, eLife introduced its first computationally reproducible document, based on a research article originally published in the Reproducibility Project: Cancer Biology by Tim Errington, the Director of Research at the Center for Open Science. The interactive article is a demonstration of some of the capabilities of the initial prototype of the Reproducible Document Stack (RDS), an open-source tool stack for authoring and publishing reproducible articles developed by Substance and building on technology from Stencila and Binder. The demo also showcased eLife’s vision for the future of research articles.
The research community’s response to the article was overwhelmingly encouraging: thousands of researchers explored the paper’s in-line code block re-execution abilities by manipulating its plots, and several authors approached us directly to ask how they might publish a reproducible version of their own manuscripts.
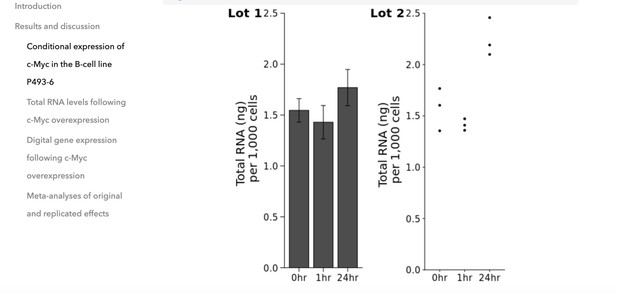
Changing the plot on the right from a bar plot to a dot plot by in-browser R-code re-execution.
Encouraged by the community interest and feedback, we have now started working on achieving a scalable implementation and service infrastructure to support the publication of reproducible articles. The goal of this next phase in the RDS project is to ship researcher-centred open-source solutions that will allow for the hosting and publication of reproducible documents, at scale, by anyone. This includes building conversion, rendering and authoring tools, and the backend infrastructure needed to execute reproducible articles in the browser.
Interoperability, modularity and openness are at the heart of the RDS’s design and development. We want authors, readers and publishers from different research communities to be able to use and interact with these tools seamlessly. RDS will continue to be developed open-source, and we strive to update and engage the community at all stages of development. Our first priority will be enabling interoperability with existing authoring tools for the Jupyter and R Markdown communities.
Interoperability, modularity and openness are at the heart of the RDS’s design and development.
As a first step, eLife aims to publish reproducible articles as companions of already accepted papers. We will endeavour to accept submissions of reproducible manuscripts in the form of DAR files by the end of 2019.
We outline the key areas of innovation of this next phase of RDS development below.
Format converters for reproducible documents
Despite the fact that most research articles are still written in Word, an increasing number of scientists are moving to more reproducible formats such as Jupyter Notebooks and R Markdown. The new DAR format for reproducible documents finally offers a way to take this reproducibility through to publication; but without publisher support or any easy way to convert between formats, researchers fall back to converting their work to Word or PDFs before submitting to publishers, thereby losing reproducibility.
Interoperability is key when working with scientists across disciplines. We want it to be easy for a scientist to create a reproducible document from multiple starting points. Format converters will allow researchers using Jupyter Notebooks and R Markdown to submit reproducible articles to eLife and have them converted to the DAR format, without losing the reproducible elements.
We want it to be easy for a scientist to create a reproducible document from multiple starting points.
Reproducible execution services and supporting tools development
To publish reproducible documents, publishers and researchers need reliable and performant reproducible execution environments as the backend for running live-code elements. Stencila Hub is being built to provide such environments, and for the next phase of RDS development, additional functionality for the Hub will be specifically designed by Stencila for the publisher use case. The aim is to build a robust and scalable software and personnel infrastructure for the provision of execution services in support of the RDS, as well as to enhance the overall user experience. Stencila will provide the reproducible execution services to eLife, and subsequently to any publishers interested in making RDS publications available to their customers, through a Service Level Agreement.
We greatly value existing technology and the ongoing efforts of the open-source community, but at the same time, it is important that we encourage innovation and support the exploration of new approaches and solutions. We aim to maximise our reuse of existing open-source software, such as Jupyter kernels, Kubernetes, Jupyter Hub and Binder, to deliver execution services robustly and cost-effectively. Stencila’s short-term plan is to deploy its own BinderHub instance for the hosting of eLife’s reproducible articles; if alternative implementations are warranted, we will conduct thorough research and comparisons on user experience and performance, and deliver any alternative implementations as complements to existing technologies.
We also believe that there is still considerable room for innovation in the arena of efficient and scalable reproducible computation, and that a joint effort between the eLife and Stencila communities will facilitate the exploration of more robust publishing infrastructures. For example, Docker images, which are currently used to build reproducible research environments, can be very large. Stencila will therefore explore solutions to optimise the size of Docker images to the bare minimum required by a reproducible document, with the end goal of producing well-documented, tested and modular tools that will enable more reproducible and efficient execution environments. We hope to contribute these open tools and ideas towards other publishing and reproducibility pipelines beyond eLife and the RDS project.
File format specification for portable reproducible documents
Texture is an open-source editing software designed and developed by Substance specifically to edit and annotate scientific content. It uses the DAR format, which is based on a stricter form of JATS-XML. To improve the portability of reproducible documents, Substance will extend the DAR file format specification to natively support R Markdown’s inline code cells, giving the DAR file format greater interoperability with mainstream tools for computational reproducibility.
A Stencila plugin for Texture
Substance will also work on improving the Texture authoring client by extending it with a new extension architecture that will allow plugins, such as one encompassing Stencila’s code-authoring functionality, to be added into the Texture client. This will ensure Texture can be maintained as primarily an XML editor, for use in the authoring and production of traditional manuscripts, but also extended as needed via separately maintained plugins to encompass new functionality such as Reproducible Document authoring and execution. A reference Stencila plugin will also be created as the first to use the new plugin architecture.
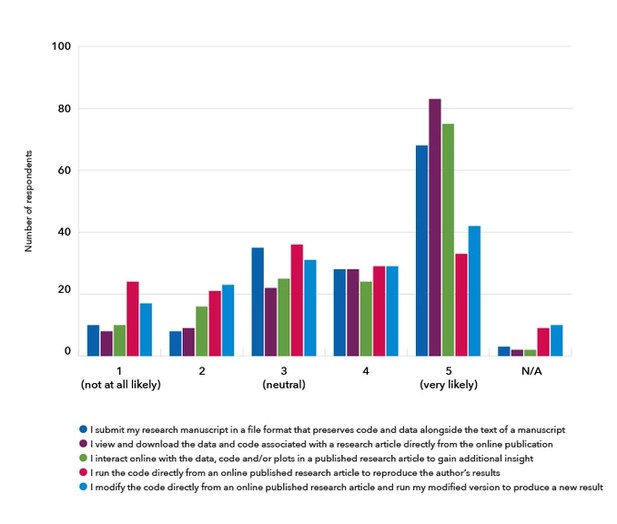
In 2017, researchers familiar with reproducible document tools told us they were interested in being able to share and read research articles with features that support better code and data sharing as well as greater interactivity and executability.
Authoring and publishing a reproducible article: the future workflow
Once the technical work on the next phase of the RDS project is complete, we envision the following workflow for authoring and publishing reproducible articles:
- Authoring. The author will follow provided guidelines for how to write reproducible articles as R Markdown, Jupyter Notebook or DAR documents. These guidelines will include how to add the necessary metadata to each of these formats.
- Uploading. The author uploads their article, and necessary data and code files, to a “project” on Stencila Hub. Eventually this step may be folded into a publisher’s submission workflow.
- Building. A compact and efficient reproducible execution environment is built for the article based on the software packages used in it. These tools also create a manifest of the software dependencies of the article, thereby providing data for software citations.
- Verification. The article is executed headlessly on the Stencila Hub within the reproducible execution environment to verify that it is indeed reproducible.
- Conversion. Once the article has been verified as being reproducible, the author presses a “Create DAR” button (when not already using the format) to export their article to DAR ready for eLife’s production team. Any issues with conversion would be reported to the user so that they could make corrections to their original document.
- Publication. A reproducible companion version of the article can be made available to readers via two mechanisms:
- The article is converted to HTML and served from the Stencila Hub. It is progressively enhanced using Javascript to make the reproducible elements live by connecting to the execution environment built for the article (also hosted on the Hub).
- The DAR is rendered by Javascript within the browser using a Texture Reader interface that is hosted by eLife and which connects to the execution environment built for the article and hosted on the Hub (this is the setup used in the demo).
Get involved
Since the release of eLife’s first reproducible article, we have been actively collecting feedback from both the research and the open-source community, and this has been crucial to shaping the development of the RDS.
We strive to continue this for the next phase of the project, which is expected to last for about a year. We hope to post frequent updates on the milestones here on eLife Labs, and we welcome your input and feedback throughout, whether about the concept or specific technical elements. Please annotate publicly on this and future blog posts.
If you'd like to know more about the RDS project, or are a researcher or developer wishing to contribute to the project, here are some key resources to get you started:
- Stencila Desktop installation instructions and code repository.
- Texture repository, the open-source manuscript editor that Stencila Desktop is based on.
- See the guide on the Stencila Converters and the relevant repository.
- Join Stencila’s chat channel and Community Forum.
- Sign up for the RDS community newsletter.
- Join Stencila’s upcoming community call on June 3 (1pm PST), and eLife’s open-source community call on June 24.
We will discuss the RDS project and reproducibility in general at various conferences over the next two months:
- Cascadia R Conf – June 8, Bellevue, US (Stencila).
- Writing reproducible & executable scientific papers with R & Python: a hands-on workshop – June 10–11, Seattle, US (Stencila).
- “Hack and Yack” session at the British Library – June 17, London, UK (eLife).
- OAI11 UNIGE-CERN Workshop on Innovations in Scholarly Communications – June 19–21, Geneva, Switzerland (eLife).
- CarpentryConnect 2019 – June 25–27, Manchester, UK (eLife).
- ISCB/ECCB/BOSC 2019 – July 21–25, Basel, Switzerland (eLife).
If you have specific questions or comments, you can also email us at innovation [at] elifesciences [dot] org, or interact with us on Twitter @eLifeInnovation.
#
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
Are you interested in contributing to open-source projects like the Reproducible Document Stack to drive forward open science? Applications are open until June 2 for the eLife Innovation Sprint in September 2019.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




